Hello Everyone,
In some of earlier posts I have talked about the classification of microbial world and what a Microbial Taxonomist do to characterize them? So, I thought why not to expand it and explain it in full detail, the identification and classification system of these microbes. Bacterial taxonomy is not something which excites the new researchers or scientists to work on until the development of techniques involving the sequencing of rRNA, the complementary DNA and the role of these genes in the phylogeny system of microbial species and all organism came into picture. All these new techniques motivates us for the phylogeny and the taxonomy of all species, where bacterial taxonomy could also be given a space in phylogeny.
Polyphasic Taxonomy
Polyphasic Taxonomy came into picture 25 years ago, which considered the genotypic, phenotypic and phylogenetic approach for the bacterial taxonomy. Colwell gave the term “Polyphasic Taxonomy” and used it for the presentation of levels in the taxa. Microbial systematics or taxonomy are generally divided into three parts:
- Classification (On the basis of similarity organisms are arranged in different taxa)
- Nomenclature (Giving name to the defined taxa)
- Identification (determines the unknown or not reported organism belong to which of the taxa)
Apart from these traditional biosystematics, two additional area are needed to define the taxonomy in the advanced system: phylogeny and modern genetics. A species of a same genus can be defined to another or different species only if the DNA percentage of the relatedness between the new species and the type strain is below 70%. The type strain usually considered to be the species which was first belong to that genus and considered to be new species and after that all are considered to be subspecies.
Information from the Bacterial Polyphasic Taxonomy
Polyphasic taxonomy generally involves the characterization study of genotype, phenotype and phylogenetic information. The genotypic information involves the data came from the nucleic acid (DNA and RNA) in cell, while the phenotypic data deals with the different proteins they produced and their function and different chemotaxonomic markers and more. I’ll try to just give a brief of all the different types of characterization studies involved in the process of characterizing a new species.
 G+C content%
G+C content% DNA-DNA hybridisation
DNA-DNA hybridisation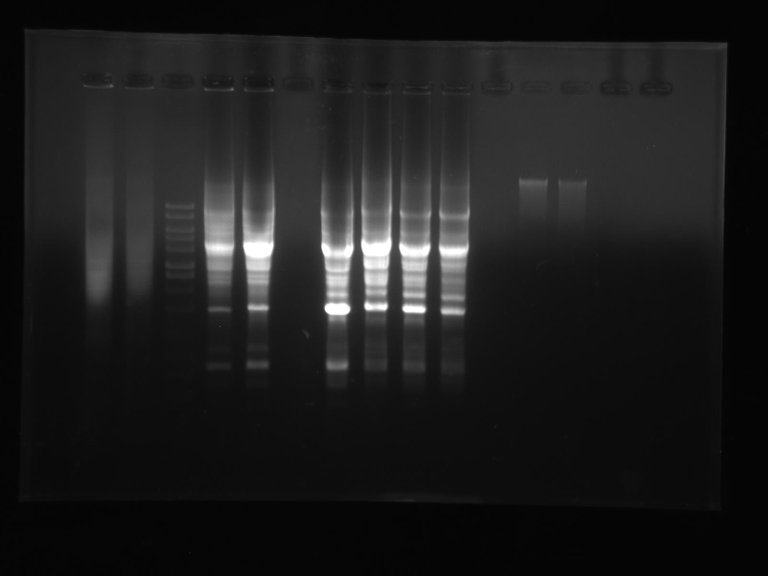 DNA based typing
DNA based typing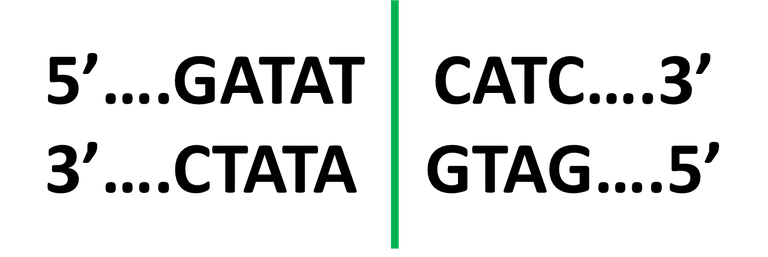 Restriction endonuclease
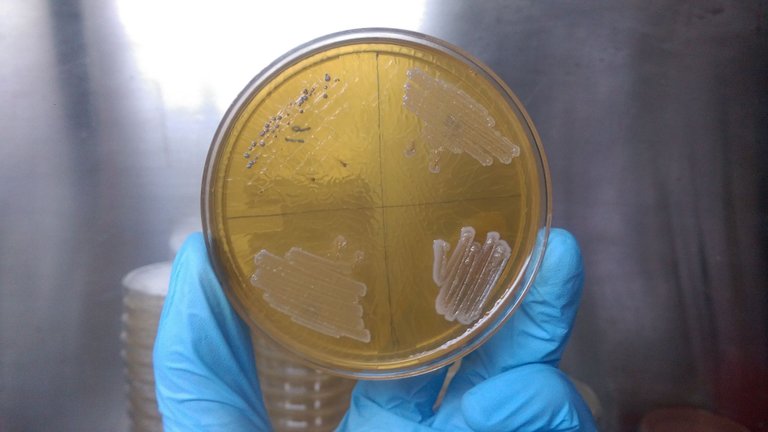
Restriction endonuclease Bacterial Colony Morphology
Bacterial Colony MorphologyThe physiological characteristics involve the growth of bacteria in different temperature condition, different pH, different salt concentration and growth in aerobic and anaerobic condition, growth in presence of various substance like antibiotics. All these condition will group them according to the growth condition and that helps in categorizing them in the phylogeny. Growth at different temperature will group them in thermophile (hot environment), Psychrophilic species (cold environment) and Mesophilic sp. (normal environment) accordingly. The biochemical characterization generally characterize the bacterial group on the basis of protein and enzyme they produces and on several reactions they produces reaction byproducts which confirms the presence of that bacterial species. There are several other tests which are used to categorize the bacterial species in a particular taxa like determining the cell wall composition, this might be a tedious process but also proposes a rapid screening of a large population.

Chemotaxonomy came into picture few years back and considered to be a milestone in the process of bacterial classification, as the information it gives can also alone give the classification of a bacterial species. The term “Chemotaxonomy” refers the chemical composition and its study for the classification and placing a bacterial species in its taxa. Cellular fatty acid present in the bacterial cell membrane vary in different species, a variety of lipids can be found in the membrane and on the basis of these lipids they can be classed into different groups. Phosphatidylethanolaminie (PE), phosphatidylcholine (PC), phosphatidylglycerol (PG), sphinogolipids and dimeric subunits of these and many more of these lipids can be found in the bacterial species.
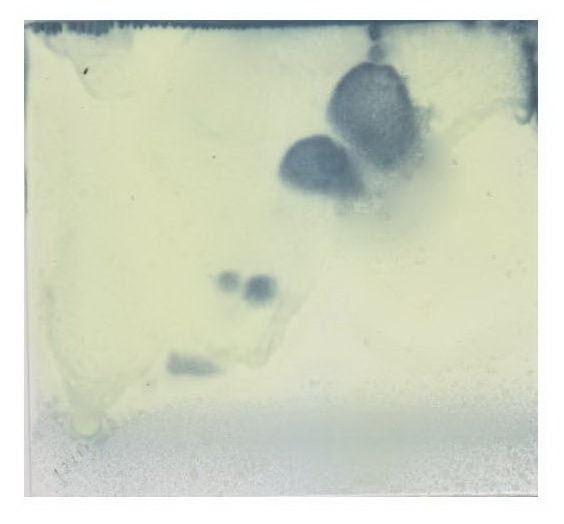 Polar Lipid Profile
Polar Lipid ProfileIsoprenoid quinones and Polyamines are also used to differentiate the bacterial population on the basis of their distribution in the cell membrane. Isoprenoid are generally involved in the electron transport chain, oxidative phosphorylation and active transport in the cell. Phylloquinone and menaquinone are the subgroup which used to differentiate the two bacterial species and they are quantified by HPLC. The role of the polyamines in the bacterial cell is not clearly known till now but can be used to differentiate between bacterial species, these polyamines are also been determined by the gas chromatography and high-performance liquid chromatography.
Evolution in the bacterial taxonomy
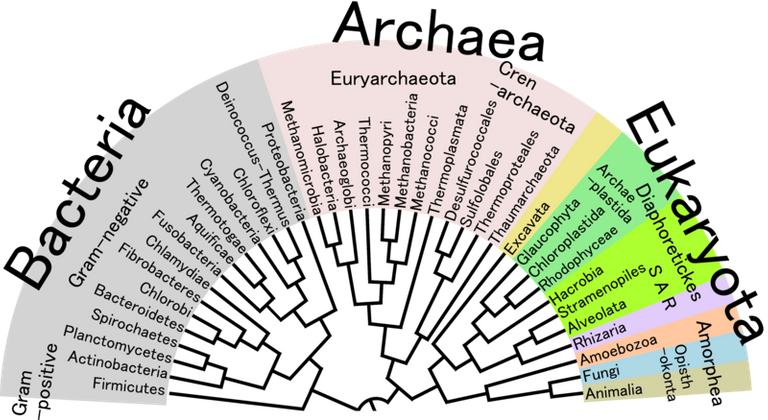
With all these advancements in the taxonomy method and the involvement of genome to the study not only evolved the whole procedure but also made it interesting for the coming generation. As this area was thought to be dull and boring, these advances has given it a new life and helps it in the evolution. Based on all these characteristics the phylogenetics of the new species can be made, which utilizes several algorithm to build a tree based on the rRNA sequence and also support the other characterization results. The phylogenetic tree shows the similarity index with the closely related subspecies and subfamilies.
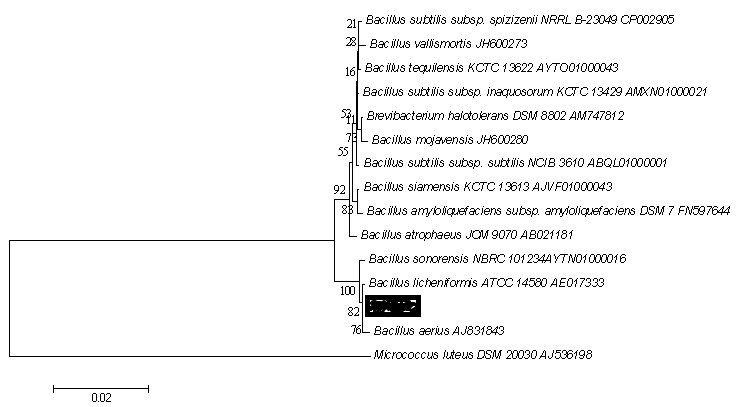
Phylogenetic Tree
The whole process of the polyphasic taxonomy is a wide area and I have described it here in brief and some of the important techniques and methods for the characterization will be shared in future just keep reading them.
Images which are not sourced are taken by me and do not violates any copyright.
References
Prakash et. al., 2007 Indian Journal of Microbiology, 47(2);98-108.
Vogel et. al., 2000 Int. Jour. Syst. Microb., 50;1133-1142.
Gillis et.al., 2015 Bergey’s Manual.
Moore et.al., 2010 Res Microbiol. 161(6);430-438.
Cowan et.al., 1965 J. gen. Microbiol. 39;148-158.
Vandamme et.al., 2015 Clinical Microbiology (Book)
Chang Cho et.at., Appl. Environ. Microbiol. 67(8);3677-3682.
Chibucos et.al., 2006 Appl. Environ. Microbiol. 72(5);3350-3356.
Donelli et. al., 2013 Microb. Ecol. Health Dis. 24(2).
Vandamme et. al., 1996 Microbiol. Rev. 60(2);407-438.
Hope you like the read, please upvote and follow my page for reading more of such articles on microbiology.
Thank You
Good one friend
Is the manual DNA-DNA hybridization method is still in fashion, or it gives the better result in comparison to the digital DNA-DNA hybridization?
The better one is Digital DNA-DNA hybridization only but its bit expensive as compared to the manual one, in digital we need to have the whole genome of all the species. Let's say you need to analyse among 3-4 closely related species than you need to sequence all the 4 species than only and its bit expensive. But the result form the digital hybridization can not be compared to the manual one as it is very accurate.
where as the manual hybridization needs the experiment in triplicates to give the real picture.
Thanks for the comment.
Your Post Has Been Featured on @Resteemable!
Feature any Steemit post using resteemit.com!
How It Works:
1. Take Any Steemit URL
2. Erase
https://3. Type
reGet Featured Instantly & Featured Posts are voted every 2.4hrs
Join the Curation Team Here | Vote Resteemable for Witness